Sidebar
**This is an old revision of the document!**
Table of Contents
Slurm basics
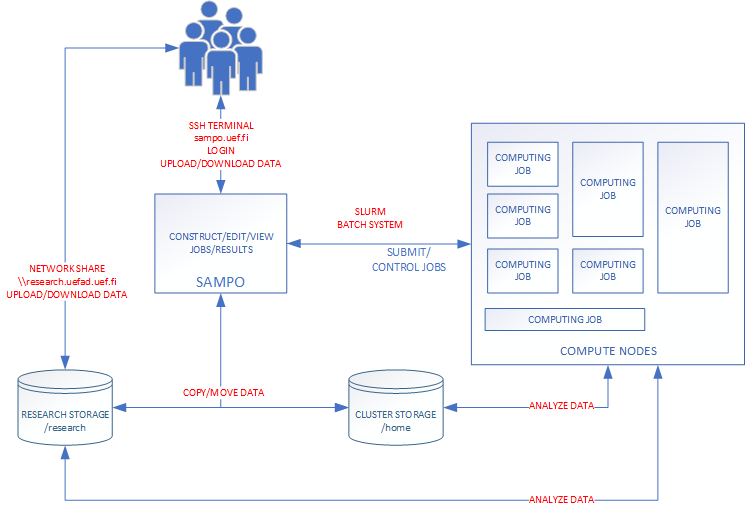
Slurm Workload manager is an open source, fault-tolerant, and highly scalable cluster management and job scheduling system for large and small Linux clusters. As a cluster workload manager, Slurm has three key functions.
- It allocates exclusive and/or non-exclusive access to resources (compute nodes) to users for some duration of time so they can perform work.
- It provides a framework for starting, executing, and monitoring work (normally a parallel job) on the set of allocated nodes.
- It arbitrates contention for resources by managing a queue of pending work.
- Optional plugins can be used for accounting, advanced reservation, gang scheduling (time sharing for parallel jobs), backfill scheduling, topology optimized resource selection, resource limits by user or bank account, and sophisticated multifactor job prioritization algorithms.
Bioinformatics Center uses unmodified version of Slurm on sampo.uef.fi computing cluster. This guarantees that the most of the tutorials and guides found from the Internet are applicable as-is. The most obvious starting place to search for usage information is documentation section of the Slurm own website Slurm Workload Manager.
Example
Example MATLAB code (matlab.m)
% Creates a 10x10 Magic square M = magic(10); M
Example script (submit.sbatch)
#!/bin/bash module load matlab/R2018b # load modules matlab -nodisplay < matlab.m # Execute the script
User can submit the job to the compute queue with the sbatch command.
sbatch submit.sbatch
sbatch command submits the job to the job queue and executes the bash script. Output of the job can be found from the local output file.
less slurm-jobid.out
Slurm job queue
User can monitor the progress of the job with the squeue command. JOBID is provided by the sbatch commmand when the job is submitted.
squeue -j jobid
Scontrol - View or modify Slurm configuration and state.
Scontrol command gives some information about the job, queue (partition) or the compute nodes. This tool can also modify various parameters of submitted job (runtime for example).
List all compute nodes
scontrol show node
List all compute nodes
scontrol show node
List all queues/partitions
scontrol show partition
List information of the given jobid
scontrol show job jobid
Extend runtime of given jobid
scontrol update jobid=<job_id> TimeLimit=<new_timelimit>
Slurm job effiency report (seff)
Seff command will give the report of the completed job on how much resources it consumed. The reported information are CPU wall time, job runtime and memory usage.
seff jobid